Gene Set: D4 : CTD - Gossypol
| Collection | D4 : CTD | ||||||||||||||||
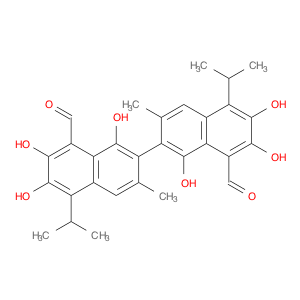
| Chemical Name | Gossypol ( From PubChem : ���G���O���S���S���Y���P���O���L ) | ||||||||||||||||
|
|||||||||||||||||
|
|||||||||||||||||
| Structure |
|
||||||||||||||||
| InChI | ���I���n���C���h���I���=���1���S���/���C���3���0���H���3���0���O���8���/���c���1���-���1���1���(���2���)���1���9���-���1���5���-���7���-���1���3���(���5���)���2���1���(���2���7���(���3���5���)���2���3���(���1���5���)���1���7���(���9���-���3���1���)���2���5���(���3���3���)���2���9���(���1���9���)���3���7���)���2���2���-���1���4���(���6���)���8���-���1���6���-���2���0���(���1���2���(���3���)���4���)���3���0���(���3���8���)���2���6���(���3���4���)���1���8���(���1���0���-���3���2���)���2���4���(���1���6���)���2���8���(���2���2���)���3���6���/���h���7���-���1���2���,���3���3���-���3���8���H���,���1���-���6���H���3 | ||||||||||||||||
| InChIKey | ���Q���B���K���S���W���R���V���V���C���F���F���D���O���T���-���U���H���F���F���F���A���O���Y���S���A���-���N | ||||||||||||||||
| Links |
|
||||||||||||||||
Gene (68 / 193) ⊖ Less |
|
||||||||||||||||
| Download gene sets | gmt, text, Detailed text | ||||||||||||||||